Journal of Cancer Research & Therapy
An International Peer-Reviewed Open Access Journal
ISSN 2052-4994


- Download PDF
- |
- Download Citation
- |
- Email a Colleague
- |
- Share:
-
- Tweet
-

Journal of Cancer Research & Therapy
Volume 7, Issue 1, February 2019, Pages 1–8
HypothesisOpen Access
HTLV-1 can be involved in acceleration of different nonhematological cancers
- 1 Moores Cancer Center, University of California, San Diego. 9500 Gilman Drive, La Jolla, California 92093, USA
- 2 San Diego Supercomputer Center, University of California, San Diego. 9500 Gilman Drive, La Jolla, California 92093, USA
- 3 Department of Neurosciences, University of California, San Diego. 9500 Gilman Drive, La Jolla, California 92093, USA
- 4 REHS program, SDSC, University of California, San Diego. 9500 Gilman Drive, La Jolla, California 92093, USA
- 5 CureMatch Inc. San Diego, 6440 Lusk Blvd Suite D206, San Diego, California 92121, USA
*Corresponding author: Igor Tsigelny, University of California, San Diego, 9500 Gilman Drive, Mail Stop 0505, La Jolla, CA 92093-0505, USA. Email: itsigeln@ucsd.edu. CureMatch Inc., 6440 Lusk Blvd, Suite D206, San Diego, CA 92121, USA. Email: itsigelny@curematch.com
Received 3 January 2019 Accepted 30 January 2019 Published 5 February 2019
DOI: http://dx.doi.org/10.14312/2052-4994.2019-1
Copyright: © 2019 Kouznetsova VL, et al. Published by NobleResearch Publishers. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
AbstractTop
In the past two decades it has been found that viral proteins might contribute to activation of numerous pathways involved in different types of cancer. Human T-lymphotropic virus 1 (HTLV-1), well known as a cause of T-cell leukemia, was found in patients having different types of cancers. Initiation and development of cancers is usually a multi-year multistep process that starts with initiation of the first somatic mutation and the then the next set of steps with the occurrence of an increasing number of somatic mutations with specific times for each one. Here we suggest a hypothesis: If HTLV-1 virus can infect the nonhematological cell, its viral proteins can work as the agents activating the general cancer-related pathways. This way Tax HTLV-1 protein (and/or other viral proteins) can serve in the similar way as the somatic mutation of specified genes and the presence of Tax can affect the functionality of several pathways that are not yet activated by the timeline of the stages of a “normal” cancer development. For example, the pathways PI3K, p53, TGF-β can be correspondingly be activated and inhibited by Tax before their functional change would appear by the timeline of a “normal” cancer development. The HTLV-1 infection-related KEGG pathway was found when the sets of the most frequently mutated genes for several cancers were analyzed by the DAVID program program (that selects the most probable pathways for a set of selected genes). These findings support the hypotheses that HTLV-1 infection can be a common factor in the development of very different cancers, thus warranting further exploration.
Keywords: cancer development; HTLV-1; HTLV-1 infection pathway; Tax protein; viral proteins
IntroductionTop
Initiation of cancers
Initiation and development of cancers is usually a multi-year multistep process that starts with initiation of the first somatic mutation and then occurrence of the next somatic mutations. As pointed by Harris and colleagues the rodent cells need at least two genetic aberrations before transformation to cancerous cells and human cells need more than that [1, 2]. On example of colorectal cancer (CRC), Jones and coauthors show that the initiation of cancer starts with a mutation of a gene inactivating the adenomatous polyposis coli gene (APC)/β-catenin pathway [3]. Then consequently the mutations affecting KRAS/BRAF, TGF-β, PIK3CA, TP53, and other cancer-related pathways occur causing progression of the stages of cancer. This process takes more than 17 years in humans. So, a number of specific pathways have to consequently be affected to support the cancer development during a long time.
Viral proteins as factors affecting specific cancer pathways
The nature of a retrovirus is to inject genetic material into host cells, altering pathways that control proliferation, cell death, etc., resulting in various diseases including cancer. It’s believed that 15% to 20% of cancers are in some way related to viral infections [4–6]. The well-known oncogenic viruses are: HIV [7], human T-lymphotropic virus (HTLV), human papillomavirus, hepatitis B and hepatitis C viruses, Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus (KSHV), and Merkel cell polyomavirus. A role of HTLV-1 and its mammalian homologs in development of adult T-cell leukemia (ATL) is well known [8–11]. HTLV-1 was the first retrovirus to be discovered and described [12–15]. It is unique in its encoding of Tax proteins, which regulates transcriptional initiation, and Rex proteins, which regulate the export of nuclear RNA [16–20]. HTLV is known to cause adult T-cell leukemia in about 5% of those infected. Tax protein plays an important role in the leukemogenesis. Tax affects several cancer-related pathways including apoptosis pathways, in addition to DNA damage repair pathways, causing mutations to accumulate and mutated cells to continue to proliferate [21–25].
HTLV-1 often leads to immortalization or transformation of the infected cells [26, 27]. It also affects the cell cycle and genomic stability, inhibits apoptosis, and increase production of several cytokines [21, 26, 27]. In the same time recently, new evidence appears related to the significant presence of this virus and its mammalian analogs in various (other than leukemia) types of cancer [28]. In the breast cancer patients BLV DNA in mammary is found in 59%, comparing to normal women having 29% with the significant p-value [20, 29]. Another study brought even greater presence of BLV in breast cancer patients [30, 31]. It worth to note that Gillet and Willems demonstrated absence of BLV DNA in breast cancer patient’s DNA. HTLV-1 virus also had been found in the other type of cells in different nonhematological cancers [32]: (i) Endometrial adenocarcinoma in rabbits [33], (ii) Hepatocellular carcinoma (HCC) in patients with chronic HCV infection [34], (iii) Non-small cell lung cancer [35], (iv) Small cell lung cancer [36], (v) HTLV-1 is isolated from immortalized epithelial cells [31], and primary oral keratinocytes [37]. Also, it was shown that HTLV-1 increases risk of bronchioloalveolar carcinoma [38].
Materials and methodsTop
Approach overview
In our study we used several computational programs. Using COSMIC [39–41], most frequently mutated genes were elucidated. The selected genes were analyzed by the DAVID program to find corresponding signaling pathways [42–45] derived from Kyoto Encyclopedia of Genes and Genomes (KEGG) [46]. Pathways for the studied cancers were compared in Venn diagrams [47], using VENNY [48]. The MutaGene program permitted identification of cancers with similar mutational signature profile patterns [49, 50].
The COSMIC program
Somatic mutations are a well-known feature in different cancers [24]. Each cancer creates a distinguished mutational pattern that is called a cancer mutational signature [51], which can be used for analysis. After collecting the most frequently mutated genes for the selected cancers, we elucidated them using the Catalogue of Somatic Mutations in Cancer (COSMIC) program [39–41]. COSMIC is the resource for exploring the impact of somatic mutations in human cancer [39]. COSMIC contains information with related details of somatic mutations about human cancers. Version 85 released in May 2018, includes 5 621 127 coding mutations on 1 383 362 samples that are curated. COSMIC database identifies 30 mutational signatures in human cancer [39]. COSMIC provides information with mutation counts and their frequencies [41].
The DAVID program
Obtained lists of the most frequent genes in the studied cancers were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) program [42, 44, 45] to find the related signaling pathways. The tools in the DAVID program provide functional interpretation of genes derived from genomic studies [42, 44, 45]. DAVID allows to discover enriched functional-related gene groups, visualize genes on BioCarta & KEGG pathway maps, list interacting proteins, as well as to link gene–disease associations.
The KEGG database
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for analysis of high-level functions of biological systems from molecular to ecological level [46]. It allows to compare information from metabolic and genomic features to cancer, endocrine, and virus networks including signaling, transcription, and disease compartments.
The VENNY program
Obtained pathways were compared using a Venny program [48], an interactive tool for comparing lists with Venn diagrams [47]. Venn diagrams were created for all elucidated lists of pathways for each studied cancer.
The MutaGene program
For selection of cancers with similar mutation signatures we used the program MutaGene [49] – an online computational framework, including several tools for analysis of cancer context-dependent mutations [50]. The framework explores DNA context-dependent mutational patterns and elucidates somatic cancer mutagenesis, analyzes mutational profiles of cancer samples [50]. Mutational profiles are categorized based on cancer type and primary tumor sites. Currently, MutaGene includes 9450 cancer samples with 1 139 534 nonrecurring mutations and 1953 somatic mutations identified in 70 benign TCGA samples [49].
The comparison between a pair of mutational signatures is performed using the MutaGene Compare function. The comparison reveals inter-cancer similarities in terms of their mutational profiles. A similarity for two signatures is calculated from a matrix based on the following similarity measures: Chi-square test, Cosine distance, Hellinger distance, Jensen-Shannon similarity, and Kullback–Leibler divergence [522].
ResultsTop
Initiation and development of cancer is a long multistage process. It starts from a single somatic mutation activating one pathway [1-3], with new mutations appearing during quite a long time and opening the next cancer-related pathways. When a tumor is detected (sometimes 30 years after the initiation) often half of the somatic mutations are already in the tumor [1, 2]. A simplified scheme of colorectal cancer development introduced by Jones and coauthors [3] suggests that one or two pathways (APC/β-catenin, CDC4/CIN) activation can lead to the initial adenoma, with following transformation to carcinoma with activating of PI3K, and inhibiting p53 pathways [1–3]. If one elucidates the signaling cancer-related pathways affected by HTLV-1 protein Tax, one would see a striking similarity of the abovementioned CRC pathways and Tax-affected pathways (Figure 1). The main pathways Wnt, PI3K, TGF-β, p53, MAPK are the same.
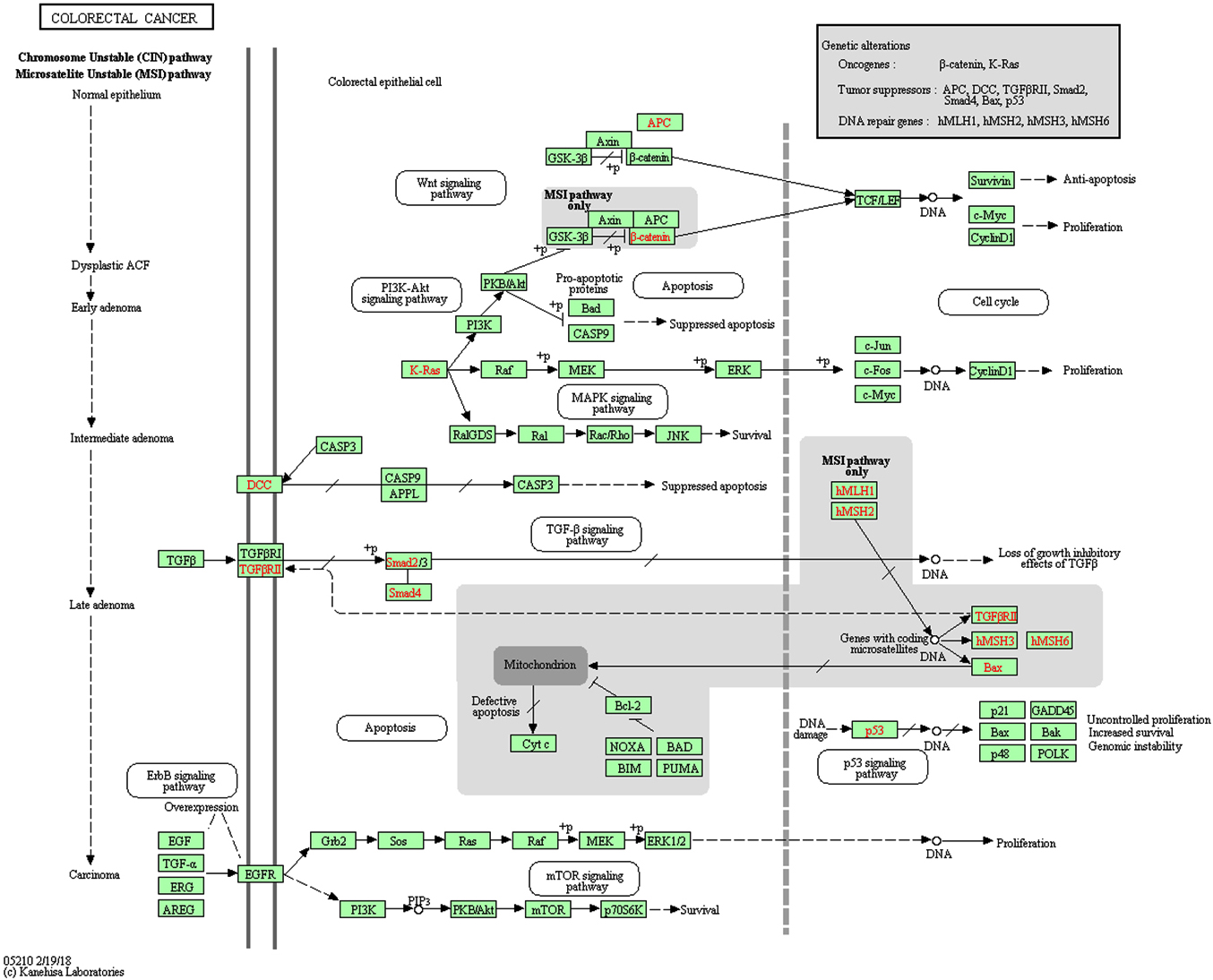
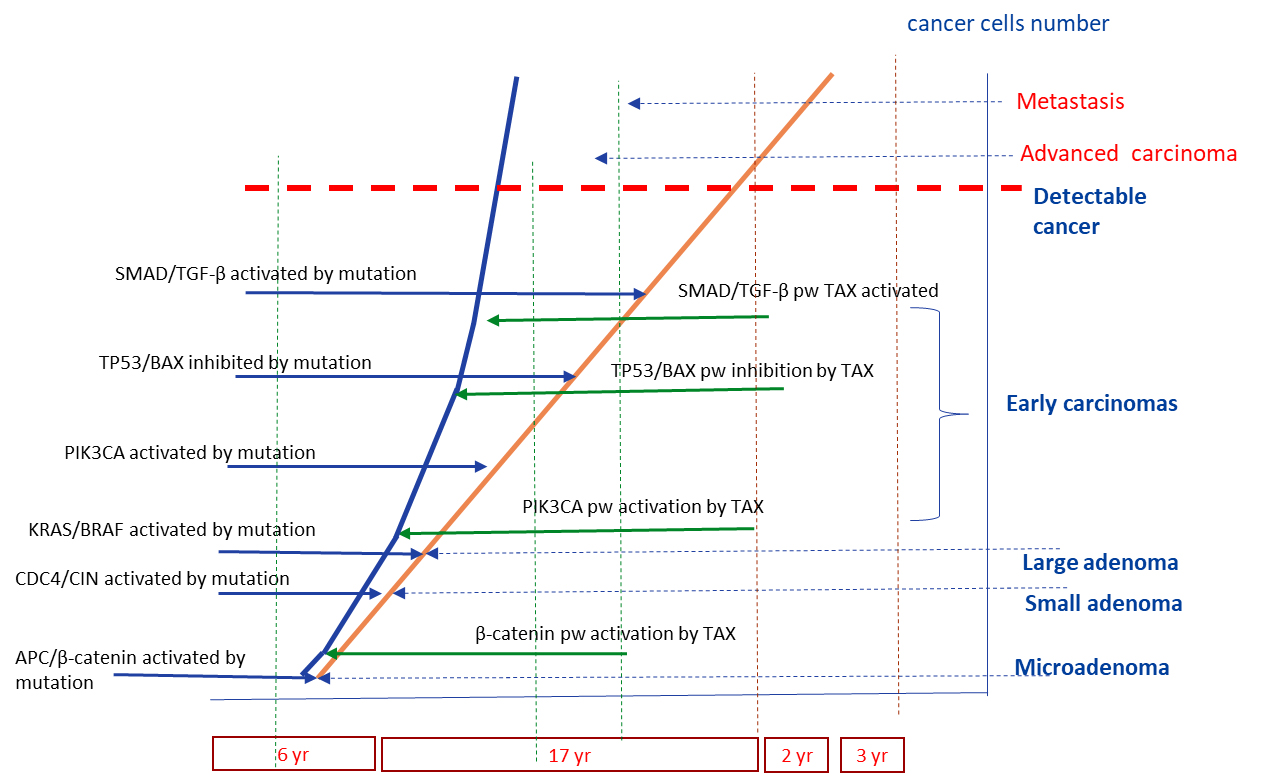
Here we want to state a hypothesis. If HTLV-1 virus can infect the nonhematological cell, its viral proteins can work as the agents activating the general cancer-related pathways. This way, the Tax HTLV-1 protein can serve in a similar way to somatic mutation of specified genes (Figure 1) and the presence of Tax can affect the functionality of several pathways that are not activated yet by the timeline of the stages of a “normal” CRC development. For example, the pathways PI3K, p53, TGF-β can be correspondingly activated and inhibited by Tax before their functional change would appear by the timeline of a “normal” cancer development. The degree of Tax impact to the cancer development can be related to a general viral load of HTLV-1. We presume that most probably Tax affects a set of pathways simultaneously. This situation may lead to shortening of the stages of cancer development because of Tax (and maybe other viral proteins) interaction with the specific pathways (Figure 2).
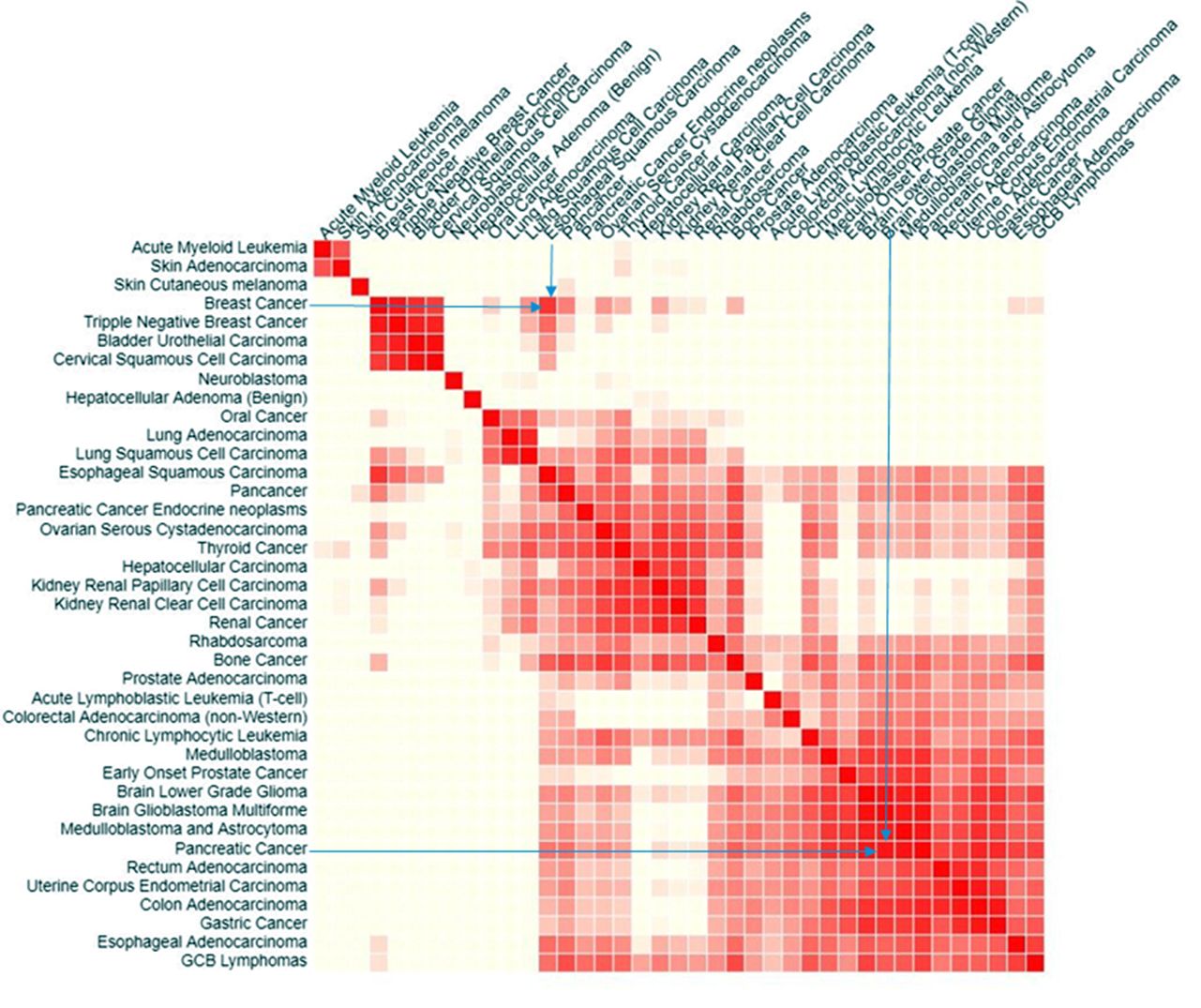
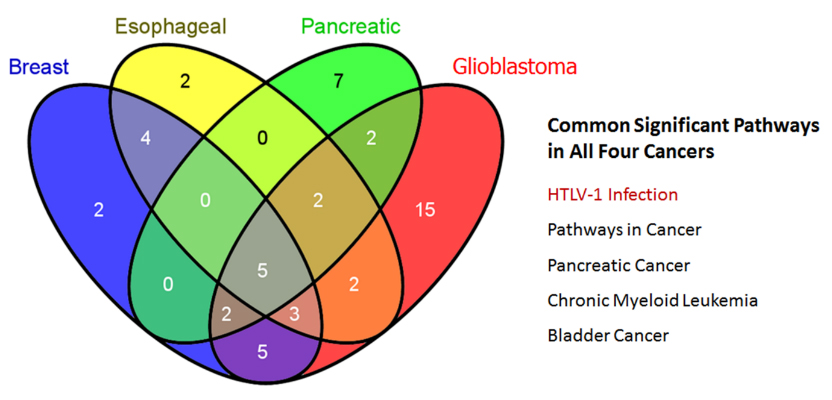
We studied pairs of significantly different cancers: breast cancer and esophageal squamous carcinoma; pancreatic cancer and glioblastoma multiforme (GBM). These pairs were selected as having a high cosine similarity of their mutational signatures (Figure 3) [53, 54]. Then we found using COSMIC, the most frequent somatic mutations in the patients having these cancers. Venn diagram (Figure 4) shows that only one common (not related directly to cancer) pathway that was found for the sets of 20 most frequently mutated genes in all abovementioned cancers is the HTLV-1 Infection pathway. Figure 5a & b, shows close-ups of parts of the HTLV-1 pathway most related to genes having maximum probability of somatic mutations in the selected four cancers. Originally this pathway in KEGG is assigned to the virus’s association with ATL and HTLV-1 associated myelopathy (HAM). Nevertheless, the genes and pathways affected are significantly related to the main cancer-related pathways in many other cancers. Many important cancer pathways containing genes most frequently mutated in the studied cancers are affected by Tax (Figure 3).
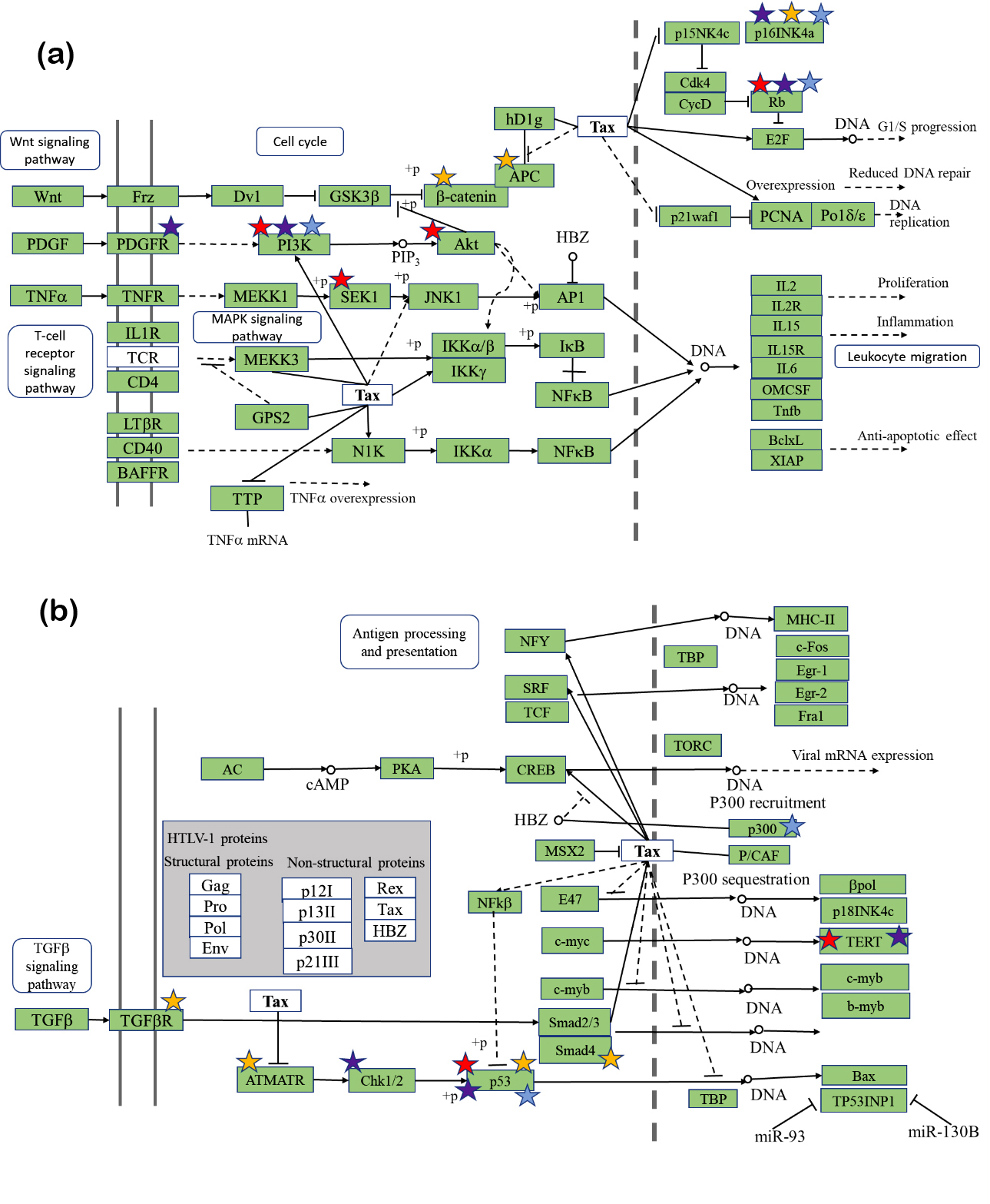
Tax is known to be a protein that interferes with signaling pathways that are related to apoptosis and cell proliferation. This list of these pathways includes among others Wnt, PDGF, TNFβ, MAPK, and TGF-β pathways. The set of these pathways shows that most probably function of Tax and other viral proteins may be not limited to leukemia and T-cells and can affect many various cancers if other types of the cells are affected. The function of this protein can include change of function of the key genes in the abovementioned pathways that supports cancer development. Tax binding to the proteins of the specific pathways may play the same role in possible activation/ inhibition of these pathways but without mutation of the genes that usually are highly mutated in mentioned cancers. In Figure 5 each protein that is encoded by a gene, which was commonly mutated in one of the four cancers is starred. Red stars correspond to breast cancer, blue stars correspond to esophageal squamous carcinoma, yellow stars correspond to pancreatic cancer, and purple stars correspond to glioblastoma. This means that if a protein is starred in all four colors, the gene encoding it was in a group of the top 20 most frequently mutated genes in all four cancers. Note that most frequently mutated genes in all four cancers are concentrated in the areas affected by the protein Tax including β-catenin, PI3K, CDK4, and MEKK1 containing pathways (Figure 6) along with p53-containing pathways and TERT (Figure 5b) [22].
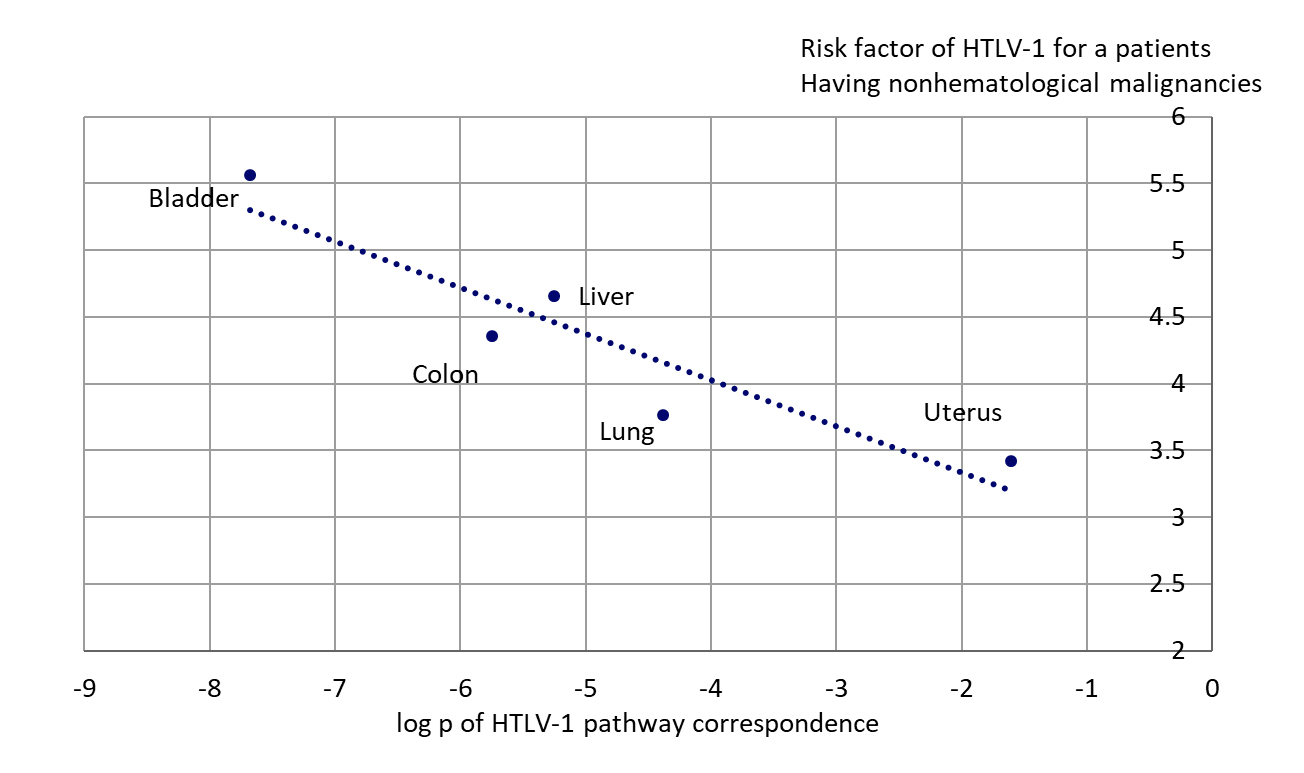
Asou and colleagues analyzed nonhematological malignancies occurring in 685 patients having HTLV-1 virus infection in Kumamoto Prefecture in southwestern Japan [56]. The patients included 397 males and 288 females with an average age 60 years. 291 patients had blood cell transfusions previously. This transfusion plays a significant role in infection by HTLV-1 virus in endemic areas (southwestern Japan is a one). During their statistical analysis, the authors calculated the HTLV-1 antibody prevalence in patients with nonhematological malignancy as a ratio of observed HTLV-1 antibody prevalence in transfused patients vs such prevalence in the healthy individuals of the Prefecture. This data was a good testing bed for our hypothesis about the possible involvement of HTLV-1 proteins in nonhematological cancers. If the hypothesis is true, we would have to see the correlation between this risk factor with the p-value of HTLV-1 pathway occurrence for the specific cancers (affected by HTLV-1). If the p-value is lowest, the most probable mutation profile of the specified cancer includes maximum of genes that may be affected by HTLV proteins. If the p-value is higher, less of the genes usually mutated in the specific cancer can be affected. Figure 6 shows dependence of the risk factor for the patients with different cancers (that had confirmed transfusion history) with log10 of p-value of HTLV-1 pathway occurrence. Data for head and neck cancer were excluded, because in general p-value for HTLV-1 pathway in the set of most frequently mutated genes of this cancer is significantly lower than for any other cancer. This case needs more study. One can see the clear dependence of HTLV-1 risk factor in patients with five different cancers with the potential corresponding mutational profiles of these cancers’ correspondence to HTLV-1 pathway probability (Figure 6). It means that the nonhematological cancers having the genes included in their possible most frequently mutated set, which are closer to the set of genes affected by Tax or other HTLV-1 proteins, would have “support” of viral proteins to accelerate these cancers development.
DiscussionTop
We showed that a number of major cancer pathways that contain genes having most frequently occurring somatic mutations can be affected directly by the HTLV 1 protein Tax. These pathways can potentially point to targets for treatment of these cancers, and the commonly mutated genes in those pathways are notable biomarkers. The HTLV 1 Infection pathway is one of the most interesting of the shared pathways [57]. HTLV 1 has been known to cause adult T-cell leukemia [14]. HTLV 1 infection is not very common in the US, but this virus is closely related to bovine leukemia virus (BLV), which has been shown to affect cows in about 84% of US dairy herds [58]. BLV and HTLV 1 both encode the regulatory proteins Tax and Rex [59, 60]. These proteins are encoded in the unique pX region, which is located between the env gene and the 3'LTR [60]. Recent studies have found antibodies in humans for BLV, suggesting that humans could be BLV-infected [61, 62]. It is not completely known yet how humans could become infected by BLV but drinking raw milk or consuming undercooked beef are possibilities. Olaya-Galan with colleagues demonstrated that raw beef and milk often contain BLV virus [63]. Given the similarities between HTLV-1 and BLV, it is possible that BLV also plays a role in the development of the other nonhematological cancers. In fact, a UC Berkeley study found that the presence of BLV DNA was associated significantly with breast cancer [27]. Further studies will have to be conducted in order to clarify the role of HTLV-1 and BLV in the development of multiple cancers. We showed that TAX protein of HTLV-1 and its mammalian analogs can affect genes that have a maximum frequency of somatic mutations in studied nonhematological cancers and are significantly involved in the development of these cancers. So, Tax can often activate or inhibit these genes without any mutations necessary and play a role of the hidden cancerogenic factor that cannot be elucidated by NGS.
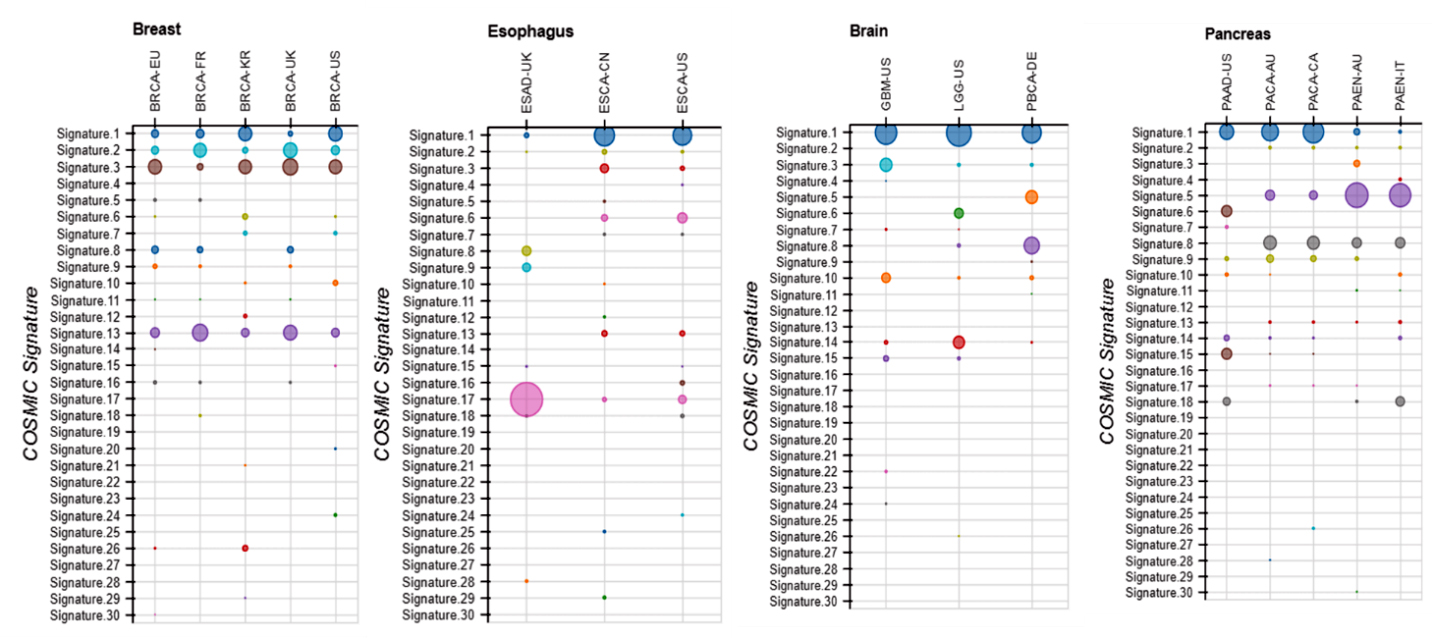
Another interesting possibility for explanation of such coherence in HTLV-1 involvement in the nonhematological cancers is a possible APOBEC-caused DNA damage. It is known that APOBEC3 proteins protect against the HTLV virus; however, they can also target mitochondrial and nuclear DNA, resulting in the development of tumors [64]. Two mutational signatures associated with APOBEC elucidated in the COSMIC database are mutational signature 2 and 13. Both are present in the breast cancer, signature 13 is present in three cancers (breast, esophagus, and pancreatic). Only GBM does not show this signature that is present in most of the cell lines for the four cancers. Signature 2 is present in all four cancers (somewhat less defined in brain and pancreatic cancers) (Figure 7). The size of the circles in Figure 7 represents the amount the mutational signature contributes to the cancer. It has been shown that APOBEC expression increases in peripheral blood mononuclear cells after HIV infection [64]. It was also shown that TFNβ is increased by HTLV-1 infection [65] that leads to an increase of APOBEC expression that can affect development of different nonhematological cancers.
Conflicts of interest
The authors declare no conflict of interest.
Acronyms and abbreviations
Akt: serine/threonine kinase 1; APC: adenomatous polyposis coli gene; APOBEC: apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; ATL: adult T-cell leukemia; ATM: ataxia telangiectasia mutated serine/threonine kinase; ATR: ataxia telangiectasia and Rad3-related serine/threonine kinase; BAX: BCL2 associated X, apoptosis regulator; BLV: bovine leukemia virus; BRAF: v-raf murine sarcoma viral oncogene homolog B; CDC4: cell division control protein 4; CDKN2A: cyclin dependent kinase inhibitor 2A; Chk1/2: checkpoint kinase 1/2; CIN: chromosomal instability CRC: colorectal cancer; CREBBPB: CREB (cAMP response element-binding) binding protein b; DAVID: database for annotation, visualization and integrated discovery; EP300: E1A binding protein p300; GBM: glioblastoma multiforme; HAM: HTLV-1 associated myelopathy; HCC: hepatocellular carcinoma; HCV: hepatitis C virus; HIV: human immunodeficiency virus ; HTLV-1: human T-lymphotropic virus 1; KEGG: Kyoto Encyclopedia of Genes and Genomes; KRAS: Kirsten rat sarcoma viral oncogene; KSHV: Kaposi's sarcoma-associated herpesvirus; MAPK: mitogen-activated protein kinase; MEKK1: mitogen-activated protein kinase kinase kinase 1 (MAP3K1); NGS: next-generation sequencing; p16INK4a: cyclin dependent kinase inhibitor 2A (CDKN2A); p53: see TP53; p300: histone acetyltransferase p300; PDGF: platelet-derived growth factors; PDGFR: platelet-derived growth factor receptor; PI3K: phosphoinositide 3-kinase; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PIK3R1: phosphoinositide 3-kinase regulatory subunit 1; PTEN: phosphatase and tensin homolog; Rb: retinoblastoma-associated protein; SEK1: dual specificity mitogen-activated protein kinase kinase (MKK4); SMAD4: Caenorhabditis elegans (Sma) genes and the Drosophila Mad (Mothers against decapentaplegic) protein 4; Rex: Tax: HTLV-1 produced proteins; TERT: telomerase reverse transcriptase; TGF-β: transforming growth factor beta; TGFβR: transforming growth factor beta receptor; TNFβ: tumor necrosis factor alpha; TP53: tumor protein 53; Wnt: wingless/integrated type family protein.
ReferencesTop
[1]Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, et al. Creation of human tumor cells with defined genetic elements. Nature. 1999; 400(6743):464–468.Article Pubmed
[2]Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000; 100(1):57–70.Article Pubmed
[3]Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008; 105(11):4283–4288.Article Pubmed
[4]zur Hausen H. Viruses in human cancers. Science. 1991; 254(5035):1167–1173.Article Pubmed
[5]Liao JB. Viruses and human cancer. Yale J Biol Med. 2006; 79(3–4):115–122.Pubmed
[6]Pickering CR, Zhang J, Neskey DM, Zhao M, Jasser SA, et al. Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin Cancer Res. 2014; 20(14):3842–3848.Article Pubmed
[7]Shiels MS, Engels EA. Increased risk of histologically-defined cancer subtypes in HIV-infected individuals: clues for possible immunosuppression-related or infectious etiology. Cancer. 2012; 118(19):4869–4876.Article Pubmed
[8]Yamagishi M, Fujikawa D, Watanabe T, Uchimaru K. HTLV-1-mediated epigenetic pathway to adult T-Cell leukemia–lymphoma. Front Microbiol. 2018; 9:1686.Article Pubmed
[9]Cook LB, Melamed A, Niederer H, Valganon M, Laydon D, et al. The role of HTLV-1 clonality, proviral structure, and genomic integration site in adult T-cell leukemia/lymphoma. Blood. 2014; 123(25):3925–3931.Article Pubmed
[10]Kataoka K, Nagata Y, Kitanaka A, Shiraish Y, Shimamura T, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015; 47(11):1304–1315.Article Pubmed
[11]Tezuka K, Xun R, Tei M, Ueno T, Tanaka M, et al. An animal model of adult T-cell leukemia: Hhumanized mice with HTLV-1-specific immunity. Blood. 2014; 123(3):346–355.Article Pubmed
[12]Gallo RC. The discovery of the first human retrovirus: HTLV-1 and HTLV-2. Retrovirology. 2005; 2:17.Article Pubmed
[13]Yoshida M. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene. 2005; 24(39):5931–5937.Article Pubmed
[14]Coffin JM. The discovery of HTLV-1, the first pathogenic human retrovirus. Proc Natl Acad Sci U S A. 2015; 112(51):15525–15529.Article Pubmed
[15]Grone M, Koch C, Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996; 218(2):316–325.Article Pubmed
[16]Ye J, Silverman L, Lairmore MD, Green PL. HTLV-1 Rex is required for viral spread and persistence in vivo but is dispensable for cellular immortalization in vitro. Blood. 2003; 102(12):3963–3969.Article Pubmed
[17]Baydoun HH, Bellon M, Nicot C. HTLV-1 Yin and Yang: Rex and p30 master regulators of viral mRNA trafficking. AIDS Rev. 2008; 10(4):195–204.Article Pubmed
[18]Bai XT, Sinha-Datta U, Ko NL, Bellon M, Nicot C. Nuclear export and expression of human T-cell leukemia virus type 1 Tax/Rex mRNA are RxRE/Rex dependent. J Virol. 2012; 86(8):4559–4565.Article Pubmed
[19]Philip S, Zahoor MA, Zhi H, Ho YK, Giam CZ. Regulation of human T-lymphotropic virus type I latency and reactivation by HBZ and Rex. PLoS Pathog. 2014; 10(4):e1004040.Article Pubmed
[20]Giam CZ, Semmes OJ. HTLV-1 infection and adult T-cell leukemia/lymphoma-A tale of two proteins: Tax and HBZ. Viruses. 2016; 8(6):pii: E161.Article Pubmed
[21]Hall WW, Fujii M. Deregulation of cell-signaling pathways in HTLV-1 infection. Oncogene. 2005; 24(39):5965–5975.Article Pubmed
[22]Hollingworth R, Grand RJ. Modulation of DNA damage and repair pathways by human tumour viruses. Viruses. 2015; 7(5):2542–2591.Article Pubmed
[23]Nicot C. HTLV-I Tax-mediated inactivation of cell cycle checkpoints and DNA repair pathways contribute to cellular transformation: "A random mutagenesis model". J Cancer Sci. 2015; 2(2).Pubmed
[24]Ryan EL, Hollingworth R, Grand RJ. Activation of the DNA damage response by RNA viruses. Biomolecules. 2016; 6(1):2.Article Pubmed
[25]Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010; 2(1):a001008.Article Pubmed
[26]Mahieux R, Gessain A. HTLV-1 and associated adult T-cell leukemia/lymphoma. Rev Clin Exp Hematol. 2003; 7(4)336–361.Pubmed
[27]Mahieux R, Gessain A. Adult T-cell leukemia/lymphoma and HTLV-1. Curr Hematol Malig Rep. 2007; 2(4):257–264.Article Pubmed
[28]Kozuru M, Uike N, Muta K, Goto T, Suehiro Y, et al. High occurrence of primary malignant neoplasms in patients with adult T-cell leukemia/lymphoma, their siblings, and their mothers. Cancer. 1996; 78(5):1119–1124.Pubmed
[29]Buehring GC, Shen HM, Jensen HM, Jin DL, Hudes M, et al. Exposure to bovine leukemia virus is associated with breast cancer: A case-control study. PLoS One. 2015; 10(9):e0134304.Article Pubmed
[30]Buehring GC, Shen H, Schwartz DA, Lawson JS. Bovine leukemia virus linked to breast cancer in Australian women and identified before breast cancer development. PLoS One. 2017; 12(6):e0179367.Article Pubmed
[31]Baltzell KA, Shen HM, Krishnamurthy S, Sison JD, Nuovo GJ, et al. Bovine leukemia virus linked to breast cancer but not coinfection with human papillomavirus: Case-control study of women in Texas. Cancer. 2018, 124(7):1342–1349.Article Pubmed
[32]Gillet NA, Willems L. Whole genome sequencing of 51 breast cancers reveals that tumors are devoid of bovine leukemia virus DNA. Retrovirology. 2016; 13(1):75.Article Pubmed
[33]Zhao TM, Bryant MA, Kindt TJ, Simpson RM. Monoclonally integrated HTLV type 1 in epithelial cancers from rabbits infected with an HTLV type 1 molecular clone. AIDS Res Hum Retroviruses. 2002; 18(4):253–258.Article Pubmed
[34]Tokunaga M, Uto H, Oda K, Tokunaga M, Mawatari S, et al. Influence of human T-lymphotropic virus type 1 coinfection on the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Gastroenterol. 2014; 49(12):1567–1577.Article Pubmed
[35]Robinson LA, Jaing CJ, Campbell CP, Magliocco A, Xiong Y, et al. Molecular evidence of viral DNA in non-small cell lung cancer and non-neoplastic lung. Br J Cancer. 2016 115(4):497–504.Article Pubmed
[36]Matsuzaki H, Hata H, Asou N, Yoshida M, Matsuno F, et al. Human T-cell leukemia virus-1-positive cell line established from a patient with small cell lung cancer. Jpn J Cancer Res. 1992; 83(5):450–457.Pubmed
[37]Dominguez MC, Enith Gonzalez N, Sanchez A, Garcia Vallejo F. Human T-Lymphotropic virus (HTLV) type I in vivo integration in oral keratinocytes. Braz J Microbiol. 2011; 42(1):310–320.Pubmed
[38]Nomori H, Mori T, Iyama K, Okamoto T, Kamakura M. Risk of bronchioloalveolar carcinoma in patients with human T-cell lymphotropic virus type 1 (HTLV-I): Case-control study results. Ann Thorac Cardiovasc Surg. 2011; 17(1):19–23.Pubmed
[39]COSMIC v86, released 14-AUG-18. Accessed on 11 January 2019: http://cancer.sanger.ac.uk/.
[40]Cancer Browser. Accessed on 11 January 2019: https://cancer.sanger.ac.uk/cosmic/browse/tissue.
[41]Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45(D1):D777–D783.Article Pubmed
[42]DAVID Bioinformatics Resources 6.8. Accessed on 14 January 2019: https://david.ncifcrf.gov/.
[43]Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, et al. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007; 8(9):R183.Article Pubmed
[44]Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37(1):1–13.Article Pubmed
[45]Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4(1):44–457.Article Pubmed
[46]KEGG: Kyoto Encyclopedia of Genes and Genomes. Accessed on 15 January 2019: https://www.genome.jp/kegg/.
[47]Venn J. On the employment of geometrical diagrams for the sensible representations of logical propositions. Proc Camb Philos Soc. 1880; 4:47–59.
[48]Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. Venny 2.1. Accessed on 16 January 2019: http://bioinfogp.cnb.csic.es/tools/venny/index.html.
[49]MutaGene: Analysis of mutations and mutational processes in cancer. Accessed on 17 January 2019: https://www.ncbi.nlm.nih.gov/projects/mutagene/ or https://www.ncbi.nlm.nih.gov/research/mutagene/
[50]Goncearenco A, Rager SL, Li M, Sang QX, Rogozin IB, et al. Exploring background mutational processes to decipher cancer genetic heterogeneity. Nucleic Acids Res. 2017; 45(W1):W514–W522.Article Pubmed
[51]Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500(7463):415–421.Article Pubmed
[52]MutaGene. Documentation. Accessed on 17 January 2019: https://www.ncbi.nlm.nih.gov/research/mutagene/help#comparison-pair.
[53]KEGG Colorectal cancer pathway. Accessed on 14 January 2019: https://www.genome.jp/kegg-bin/show_pathway?hsa05210.
[54]Huang PJ, Chiu LY, Lee CC, Yeh YM, Huang KY, et al. mSignatureDB: A database for deciphering mutational signatures in human cancers. Nucleic Acids Res. 2018; 46(D1):D964–D970.Article Pubmed
[55]KEGG Human T-cell leukemia virus 1 infection – Homo sapiens (human). Accessed on 18 January 2019: https://www.genome.jp/kegg-bin/show_pathway?hsa05166.
[56]Asou N, Kumagai T, Uekihara S, Ishii M, Sato M, et al. HTLV-1 Seroprevalence in patients with malignancy. Cancer. 1986; 58(4):903–907.Article Pubmed
[57]Gyles C. Should we be more concerned about bovine leukemia virus Can Vet J. 2016; 57(2):115–116.Pubmed
[58]Lairmore MD. Animal models of bovine leukemia virus and human T-lymphotrophic virus type-1: Insights in transmission and pathogenesis. Annu Rev Anim Biosci. 2014; 2:189–208. Article Pubmed
[59]Aida A, Murakami H, Takahashi M, Takeshima SN. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front Microbiol. 2013; 4:328.Article Pubmed
[60]Buehring GC, Shen HM, Jensen HM, Choi KY, Sun D, et al. Bovine leukemia virus DNA in human breast tissue. Emerg Infect Dis. 2014; 20(5):772–782.Article Pubmed
[61]Buehring GC, Philpott SM, Choi KY. Humans have antibodies reactive with Bovine leukemia virus. AIDS Res Hum Retroviruses. 2003; 19(12):1105–1113.Article Pubmed
[62]Wagener R, Alexandrov LB, Montesinos-Rongen M, Schlesner M, Haake A, et al. Analysis of mutational signatures in exomes from B-cell lymphoma cell lines suggest APOBEC3 family members to be involved in the pathogenesis of primary effusion lymphoma. Leukemia. 2015; 29(7):1612–1615.Article Pubmed
[63]Olaya-Galan NN, Corredor-Figuerola AP, Guzman-Garzon TC, Rios-Hernandez KS, Salas-Cardenas SP, et al. Bovine leukaemia virus DNA in fresh milk and raw beef for human consumption. Epidemiol Infect. 2017; 145(15):3125–3130.Article Pubmed
[64]Ouaguia L, Mrizak D, Renau, S, Morales O, Delhem N. Control of the inflammatory response mechanisms mediated by natural and induced regulatory T-cells in HCV-, HTLV-1-, and EBV-associated cancers. Mediators Inflamm. 2014; 2014:564296.Article Pubmed
[65]Twizere JC, Kruys V, Lefebvre L, Vanderplasschen A, Collete D, et al. Interaction of retroviral Tax oncoproteins with tristetraprolin and regulation of tumor necrosis factor-alpha expression. J Natl Cancer Inst. 2003; 95(24):1846–1859.Article Pubmed



